Artificial and Biological Intelligence Laboratory at Boston University
The DePasquale lab is located in the Department of Biomedical Engineering at Boston University. We develop mathematical models to understand how populations of neurons perform computations to produce behavior. Broadly we take two approaches. One is data-driven: we collaborate with experimental neuroscientists to develop tailored machine learning models of neural activity to identify the algorithms that drive behaviors such as decision-making or movement. Our second approach is theoretical: we construct and analyze artificial neural network models to understand how their structure gives rise to analogous computations and other functional features observed in biological neural circuits.
Research
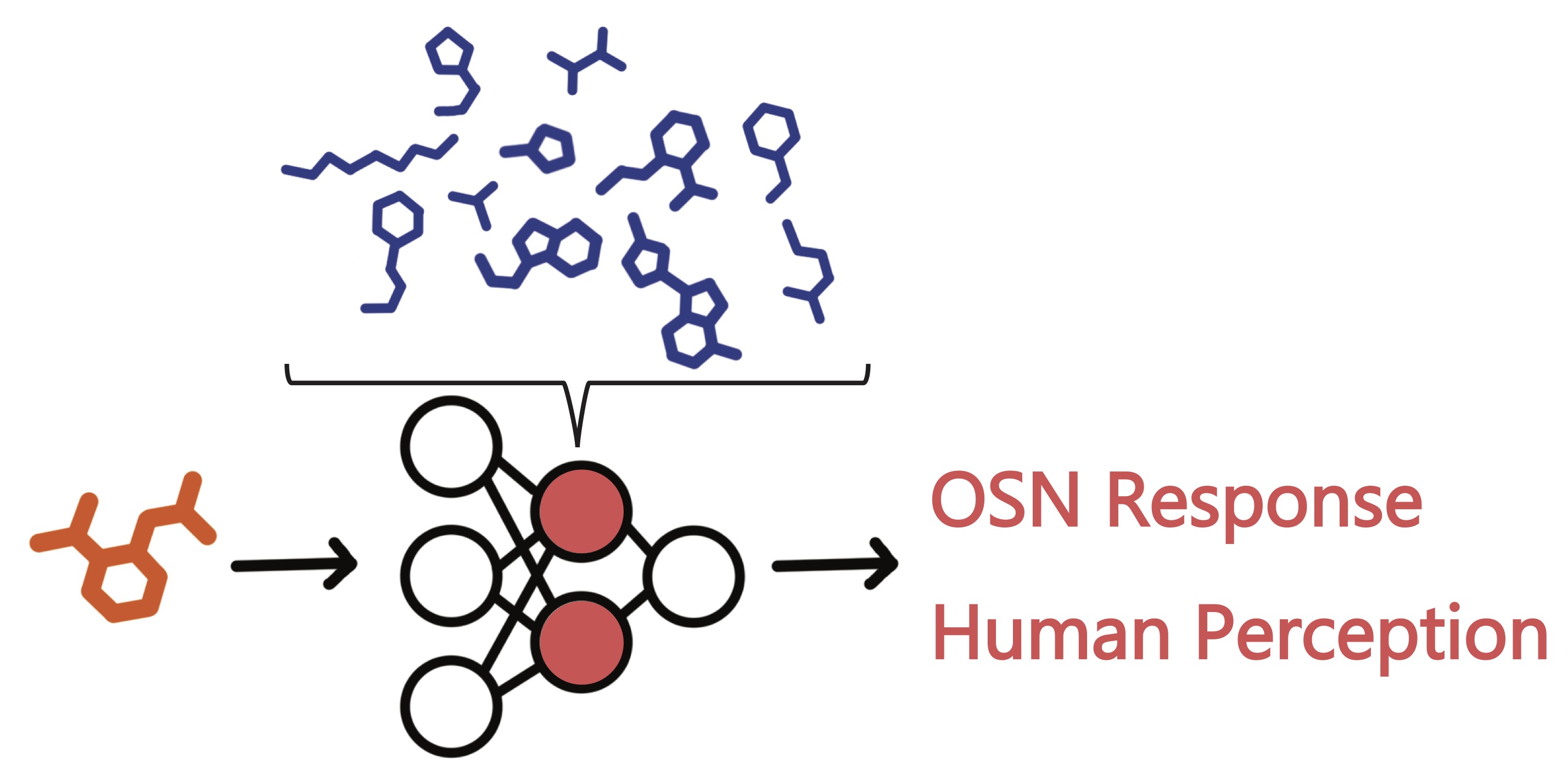
Recent advances in machine learning methods for computational chemistry have opened exciting new avenues for understanding olfaction. We are utilizing graph neural networks and self-supervised learning to identify patterns in unlabeled molecular data, generating representations of odors that are useful for predicting olfactory sensory neuron activation. These learned representations more accurately predict how individual odor molecules interact with olfactory receptors compared to existing methods.
Relevant poster: Improved odor-receptor interaction predictions via self supervised learning
McConachie, G, Duniec, E, Younger, M, DePasquale (2024)
NAIsys CSHL 2024
State-space models (SSMs) are powerful tools for modeling time series data that naturally arise in neuroscience, finance, and engineering. These models assume observations arise from a hidden latent sequence, encompassing methods like Hidden Markov Models (HMMs) and Linear Dynamical Systems (LDS). Our group is actively building an open-course software packge to implement these models in Julia called StateSpaceDynamics.jl. This modular package designed to be fast, readable, and self contained for the express purpose of fitting a plurality of SSMs, easily in Julia.
Relevant poster: StateSpaceDynamics.jl: A Julia package for probabilistic state space models (SSMs)
Senne, R., Loschinskey, Z., Loughridge, C., Fourie, J., DePasquale, B. (2024)
in prep
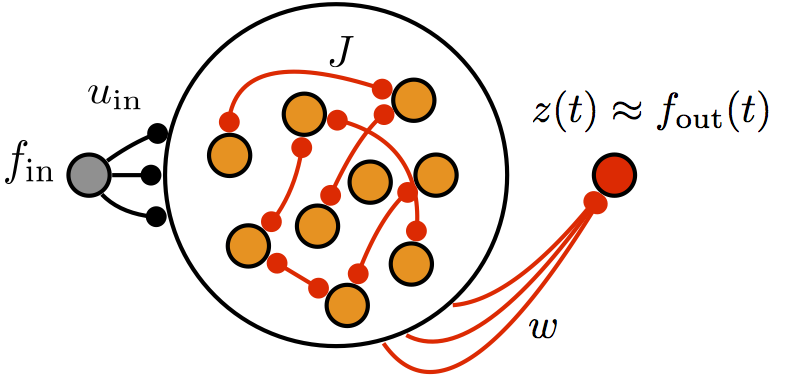
Biological neural networks compute differently than most artificial neural networks used in machine learning. For example, although real neurons communicate with spikes, reproducing this feature in artificial models has been a challenge. We develop methods for training biophysically detailed neural networks and use thse models to understand how real biologial circuits compute. Through mathematical modeling, we focus on building tighter links between biologial neural networks and more abstract artifical neural network models used in machine learning.
Relevant paper: The centrality of population-level factors to network computation is demonstrated by a versatile approach for training spiking networks
DePasquale, B, Sussillo, D., Abbott, L.F., Churchland, M.M. (2023)
in press at Neuron
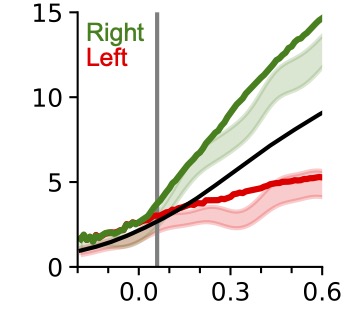
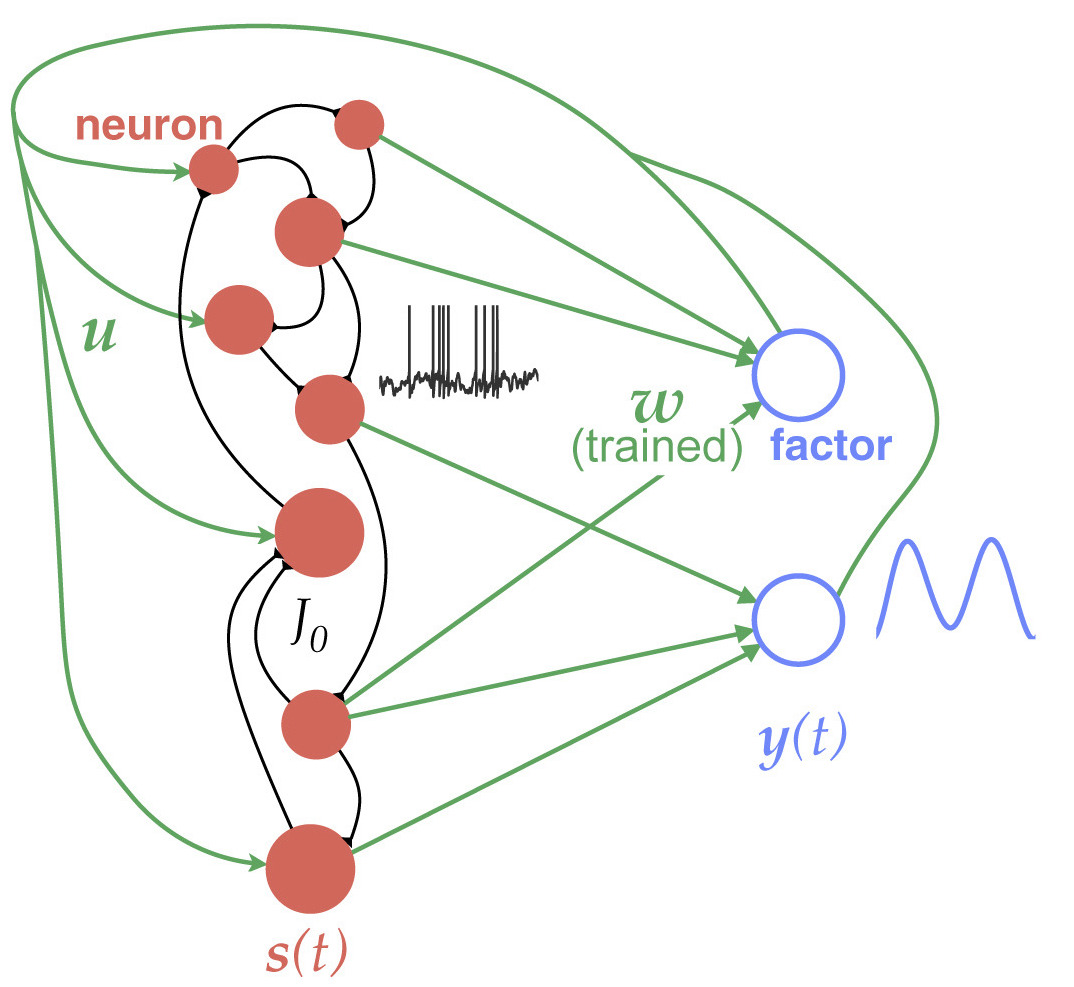
Neural recordings from behaving animals are often much too complex to link directly to an animal’s ongoing behavior. We develop machine learning models to analyze complex neural datasets to understand the algorithms that underlie different behaviors. We principally focus on the neural underpinnings of movement and decision making.
Relevant paper: Neural population dynamics underlying evidence accumulation in multiple rat brain regions
DePasquale, B., Brody, C.D., Pillow, J. (2022)
in revision at eLife
Other Recent Publications & Preprints
-
Recurrent dynamics of prefrontal cortex during context-dependent decision-making
Cohen Z, DePasquale, B., Aoi, M., Pillow, J. (2020)
bioRxiv -
Task-dependent changes in the large-scale dynamics and necessity of cortical regions
Pinto, L., Rajan, K., DePasquale, B., Thiberge, S.Y., Tank, D.W., Brody, C.D. (2019)
Neuron, 104(4), 810-824. e9 -
full-FORCE: A target-based method for training recurrent networks
code
DePasquale, B., Cueva, C.J., Rajan, K., Escola, G.S. & Abbott, L.F. (2018)
PLoS One 13(2): e0191527 -
Error-correcting dynamics in visual working memory
Panichello, M.F., DePasquale, B., Pillow, J.W. & Buschman, T.J. (2018)
Nature Communications 10, Article number: 3366 -
Building functional networks of spiking model neurons
Abbott, L.F., DePasquale, B., Memmesheimer, R.-M. (2016)
Nature Neuroscience 19:350-355
For a full list of publications, see Brian’s Google Scholar.